Population Coverage - Tutorial
1. Introduction
T cells recognize a complex between a specific major histocompatibility complex (MHC) molecule and a particular
pathogen-derived epitope. A given epitope will elicit a response only in individuals that express an MHC
molecule capable of binding that particular epitope. This phenomenon is known as denominated MHC restriction of
T cell responses. MHC molecules are extremely polymorphic and over a thousand different human MHC (HLA) alleles
are known. Selecting multiple peptides with different HLA binding specificities will afford increased coverage
of the patient population targeted by peptide-based vaccines or diagnostics. The issue of population coverage in
relation to MHC polymorphism is further complicated by the fact that different HLA types are expressed at
dramatically different frequencies in different ethnicities. Thus, without careful consideration, a vaccine or
diagnostic with ethnically biased population coverage could result.
To address the issue, this tool is implemented to calculate the fraction of individuals predicted to respond to a given epitope set on the basis of HLA genotypic frequencies and on the basis of MHC binding and/or T cell restriction data ( Bui et al. 2005). HLA allele genotypic frequencies were obtained from Allele Frequency database (http://www.allelefrequencies.net/). At present, Allele Frequency database provides allele frequencies for 115 countries and 21 different ethnicities grouped into 16 different geographical areas. In addition, the program also accepts custom populations with allele frequencies defined by users. Multiple population coverages can be simultaneously calculated and an average population coverage is generated. Since MHC class I and MHC class II restricted T cell epitopes elicit immune responses from two different T cell populations (CTL and HTL, respectively), the program provides three calculation options to accommodate different coverage modes: (1) class I separate, (2) class II separate, and (3) class I and class II combined. For each population coverage, the tool computes the following: (1) projected population coverage, (2) average number of epitope hits / HLA combinations recognized by the population, and (3) minimum number of epitope hits / HLA combinations recognized by 90% of the population (PC90).
To address the issue, this tool is implemented to calculate the fraction of individuals predicted to respond to a given epitope set on the basis of HLA genotypic frequencies and on the basis of MHC binding and/or T cell restriction data ( Bui et al. 2005). HLA allele genotypic frequencies were obtained from Allele Frequency database (http://www.allelefrequencies.net/). At present, Allele Frequency database provides allele frequencies for 115 countries and 21 different ethnicities grouped into 16 different geographical areas. In addition, the program also accepts custom populations with allele frequencies defined by users. Multiple population coverages can be simultaneously calculated and an average population coverage is generated. Since MHC class I and MHC class II restricted T cell epitopes elicit immune responses from two different T cell populations (CTL and HTL, respectively), the program provides three calculation options to accommodate different coverage modes: (1) class I separate, (2) class II separate, and (3) class I and class II combined. For each population coverage, the tool computes the following: (1) projected population coverage, (2) average number of epitope hits / HLA combinations recognized by the population, and (3) minimum number of epitope hits / HLA combinations recognized by 90% of the population (PC90).
2. Input
Parameters:
Parameter |
Description / Instruction |
Example input value |
|---|---|---|
Number of epitope(s) |
Enter number of epitope(s) in the epitope set |
50 |
Epitope |
Enter epitope name or sequence (optional) |
GILGFVFTL |
MHC restricted allele(s) |
Enter MHC alleles separated by commas or browse for alleles in the database by clicking the "Browse ..."
button |
HLA-A*02:01, HLA-A*02:02, HLA-A*02:04 |
Epitope / MHC restriction data file |
Epitope / MHC restriction data can be entered by copying data from a file. This file contains two tab delimited columns. The first column contains epitope names or sequences. The second column contains comma delimited MHC alleles. There is no header line. |
|
Population(s) / Area(s) |
Select one or more population(s) / area(s) for calculation. |
|
User population |
User defined populations can be added by clicking the "Add user population(s)"
population to copy data from a file. This file contains data in tab delimited columns. The first line is a required header line.
The first three columns are required and their headings are "MHC class", "MHC locus" and "MHC allele."
The following column headings are population names. The first column specifies MHC class (I or II). The second column specifies HLA locus
(HLA-A, HLA-B, ...). The third column specifies HLA allele ( HLA-A*01:01, HLA-A*02:01, ...). The following columns specify genotypic
frequencies (ranging from 0 to 1) of the allele for different populations. |
|
Calculation option(s) |
Check appropriate box(es) for calculation(s) of population coverage(s) based on (1) class I separate, (2) class II
separate, and/or (3) class I and class II combined |
• Format for the upload user population(s) file:
The file must contain data in tab delimited columns. The first line is a required header line. The first three columns are required and their headings are "MHC class", "MHC locus" and "MHC allele." The following column headings are population names. The first column specifies MHC class (I or II). The second column specifies HLA locus (HLA-A, HLA-B, ...). The third column specifies HLA allele ( HLA-A*01:01, HLA-A*02:01, ...). The following columns specify genotypic frequencies (ranging from 0 to 1) of the allele for different populations.
Example:
Example:
MHC Class MHC Locus MHC Allele Asian Black European-Caucasian North-America-Caucasian
I HLA-A HLA-A*01:01 0.007594 0.035415 0.167566 0.159952
I HLA-A HLA-A*02:01 0.082624 0.103242 0.258922 0.175362
I HLA-A HLA-A*02:02 0.000944 0.044661 0.007503 0.018633
...
I HLA-A HLA-A*01:01 0.007594 0.035415 0.167566 0.159952
I HLA-A HLA-A*02:01 0.082624 0.103242 0.258922 0.175362
I HLA-A HLA-A*02:02 0.000944 0.044661 0.007503 0.018633
...
• Format for the upload epitope-allele file:
File should be in simple text format. It should have only one epitope-allele combination per line. Each line should
contain a tab-separated epitope name and comma-separated allele names (example given below).
Example:
Example:
FMKAVCVEV HLA-A*02:01,HLA-A*02:02,HLA-A*02:03,HLA-A*02:06,HLA-A*68:02
FLIFFDLFLV HLA-A*02:01,HLA-A*02:02,HLA-A*02:03,HLA-A*02:06,HLA-A*68:02
GLIMVLSFL HLA-A*02:01,HLA-A*02:02,HLA-A*02:03,HLA-A*02:06,HLA-A*23:01
...
FLIFFDLFLV HLA-A*02:01,HLA-A*02:02,HLA-A*02:03,HLA-A*02:06,HLA-A*68:02
GLIMVLSFL HLA-A*02:01,HLA-A*02:02,HLA-A*02:03,HLA-A*02:06,HLA-A*23:01
...
Instructions:
- Enter number of epitopes
- Enter epitope / MHC restriction data
- Select population(s)
- Select calculation option(s)
- Click submit
Example input
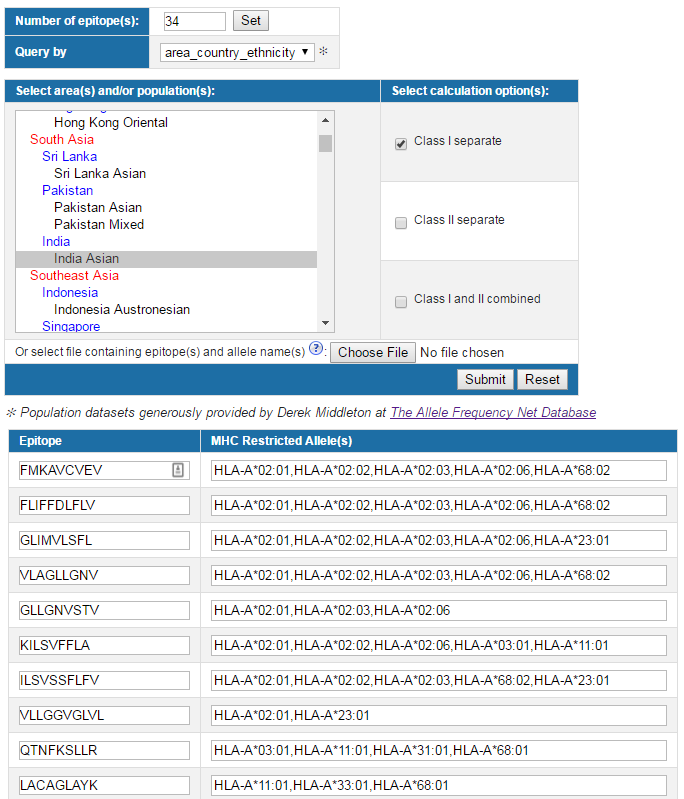
3. Output
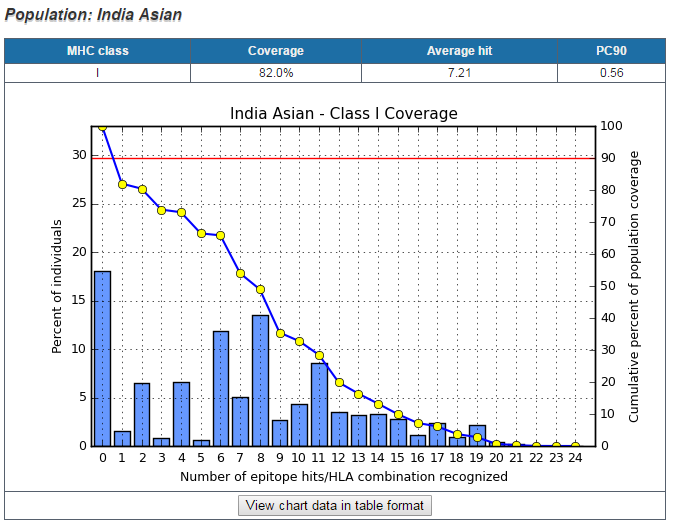
Population Information
This information was obtained from
The Allele Frequency Net Database
The HLA allele frequencies for each of the population categories were estimated as follows:
The HLA allele frequencies and associated data for different individual populations from studies around the
world were provided by allelefrequencies.net database. The populations were organized into a hierarchy based
upon geographical area, country, and ethnicity and the allele frequencies for each merged population were
estimated by combining the data from individual populations in each group. The frequencies were estimated
for alleles with resolution of 2 sets of digits (e.g. HLA-A*01:01). For this, the data for alleles with
resolution of less than 2 sets of digits were excluded (e.g. HLA-A*07, HLA-A*09 etc. which has resolution of
only one set of digits) and that for alleles with resolution of more than 2 sets of digits were combined to
2 sets of digits (e.g. HLA-A*02:01:01, HLA-A*02:01:02, HLA-A*02:01:02:01, HLA-A*02:01:02:03 etc. were
combined to HLA-A*02:01). The allele frequencies were calculated by estimating the total number of copies of
alleles (a) and the number of subjects (n) in each group of populations and using the formula, AF = a/2n.
The final set contains frequencies of 3,245 alleles (including class I and class II) for the world, 16
geographical areas, 21 ethnicities, 115 countries and ethnicities by country.
| Population Area | Population Country | Population Ethnicity |
|---|---|---|
| East Asia | ||
| East Asia | Japan | - |
| East Asia | Japan | Oriental |
| East Asia | Korea; South | - |
| East Asia | Korea; South | Oriental |
| East Asia | Mongolia | - |
| East Asia | Mongolia | Oriental |
| Northeast Asia | ||
| Northeast Asia | China | - |
| Northeast Asia | China | Oriental |
| Northeast Asia | Hong Kong | - |
| Northeast Asia | Hong Kong | Oriental |
| South Asia | ||
| South Asia | India | - |
| South Asia | India | Asian |
| South Asia | Pakistan | - |
| South Asia | Pakistan | Asian |
| South Asia | Pakistan | Mixed |
| South Asia | Sri Lanka | - |
| South Asia | Sri Lanka | Asian |
| Southeast Asia | ||
| Southeast Asia | Borneo | - |
| Southeast Asia | Borneo | Austronesian |
| Southeast Asia | Indonesia | - |
| Southeast Asia | Indonesia | Austronesian |
| Southeast Asia | Malaysia | - |
| Southeast Asia | Malaysia | Austronesian |
| Southeast Asia | Malaysia | Oriental |
| Southeast Asia | Philippines | - |
| Southeast Asia | Philippines | Austronesian |
| Southeast Asia | Singapore | - |
| Southeast Asia | Singapore | Austronesian |
| Southeast Asia | Singapore | Oriental |
| Southeast Asia | Taiwan | - |
| Southeast Asia | Taiwan | Oriental |
| Southeast Asia | Thailand | - |
| Southeast Asia | Thailand | Oriental |
| Southeast Asia | Vietnam | - |
| Southeast Asia | Vietnam | Oriental |
| Southwest Asia | ||
| Southwest Asia | Iran | - |
| Southwest Asia | Iran | Kurd |
| Southwest Asia | Iran | Persian |
| Southwest Asia | Israel | - |
| Southwest Asia | Israel | Arab |
| Southwest Asia | Israel | Jew |
| Southwest Asia | Jordan | - |
| Southwest Asia | Jordan | Arab |
| Southwest Asia | Lebanon | - |
| Southwest Asia | Lebanon | Arab |
| Southwest Asia | Lebanon | Mixed |
| Southwest Asia | Oman | - |
| Southwest Asia | Oman | Arab |
| Southwest Asia | Saudi Arabia | - |
| Southwest Asia | Saudi Arabia | Arab |
| Southwest Asia | United Arab Emirates | - |
| Southwest Asia | United Arab Emirates | Arab |
| Europe | ||
| Europe | Austria | - |
| Europe | Austria | Caucasoid |
| Europe | Belarus | - |
| Europe | Belarus | Caucasoid |
| Europe | Belgium | - |
| Europe | Belgium | Caucasoid |
| Europe | Bulgaria | - |
| Europe | Bulgaria | Caucasoid |
| Europe | Bulgaria | Other |
| Europe | Croatia | - |
| Europe | Croatia | Caucasoid |
| Europe | Czech Republic | - |
| Europe | Czech Republic | Caucasoid |
| Europe | Czech Republic | Other |
| Europe | Denmark | - |
| Europe | Denmark | Caucasoid |
| Europe | England | - |
| Europe | England | Caucasoid |
| Europe | England | Jew |
| Europe | England | Mixed |
| Europe | Finland | - |
| Europe | Finland | Caucasoid |
| Europe | France | - |
| Europe | France | Caucasoid |
| Europe | Georgia | - |
| Europe | Georgia | Caucasoid |
| Europe | Georgia | Kurd |
| Europe | Germany | - |
| Europe | Germany | Caucasoid |
| Europe | Greece | - |
| Europe | Greece | Caucasoid |
| Europe | Ireland Northern | - |
| Europe | Ireland Northern | Caucasoid |
| Europe | Ireland South | - |
| Europe | Ireland South | Caucasoid |
| Europe | Italy | - |
| Europe | Italy | Caucasoid |
| Europe | Macedonia | - |
| Europe | Macedonia | Caucasoid |
| Europe | Netherlands | - |
| Europe | Netherlands | Caucasoid |
| Europe | Norway | - |
| Europe | Norway | Caucasoid |
| Europe | Poland | - |
| Europe | Poland | Caucasoid |
| Europe | Portugal | - |
| Europe | Portugal | Caucasoid |
| Europe | Romania | - |
| Europe | Romania | Caucasoid |
| Europe | Russia | - |
| Europe | Russia | Caucasoid |
| Europe | Russia | Mixed |
| Europe | Russia | Other |
| Europe | Russia | Siberian |
| Europe | Scotland | - |
| Europe | Scotland | Caucasoid |
| Europe | Serbia | - |
| Europe | Serbia | Caucasoid |
| Europe | Slovakia | - |
| Europe | Slovakia | Caucasoid |
| Europe | Slovenia | - |
| Europe | Slovenia | Caucasoid |
| Europe | Spain | - |
| Europe | Spain | Caucasoid |
| Europe | Spain | Jew |
| Europe | Spain | Other |
| Europe | Sweden | - |
| Europe | Sweden | Caucasoid |
| Europe | Switzerland | - |
| Europe | Switzerland | Caucasoid |
| Europe | Turkey | - |
| Europe | Turkey | Caucasoid |
| Europe | Ukraine | - |
| Europe | Ukraine | Caucasoid |
| Europe | United Kingdom | - |
| Europe | United Kingdom | Caucasoid |
| Europe | Wales | - |
| Europe | Wales | Caucasoid |
| East Africa | ||
| East Africa | Kenya | - |
| East Africa | Kenya | Black |
| East Africa | Uganda | - |
| East Africa | Uganda | Black |
| East Africa | Zambia | - |
| East Africa | Zambia | Black |
| East Africa | Zimbabwe | - |
| East Africa | Zimbabwe | Black |
| West Africa | ||
| West Africa | Burkina Faso | - |
| West Africa | Burkina Faso | Black |
| West Africa | Cape Verde | - |
| West Africa | Cape Verde | Black |
| West Africa | Gambia | - |
| West Africa | Gambia | Black |
| West Africa | Ghana | - |
| West Africa | Ghana | Black |
| West Africa | Guinea-Bissau | - |
| West Africa | Guinea-Bissau | Black |
| West Africa | Ivory Coast | - |
| West Africa | Ivory Coast | Black |
| West Africa | Liberia | - |
| West Africa | Liberia | Black |
| West Africa | Nigeria | - |
| West Africa | Nigeria | Black |
| West Africa | Senegal | - |
| West Africa | Senegal | Black |
| Central Africa | ||
| Central Africa | Cameroon | - |
| Central Africa | Cameroon | Black |
| Central Africa | Central African Republic | - |
| Central Africa | Central African Republic | Black |
| Central Africa | Congo | - |
| Central Africa | Congo | Black |
| Central Africa | Equatorial Guinea | - |
| Central Africa | Equatorial Guinea | Black |
| Central Africa | Gabon | - |
| Central Africa | Gabon | Black |
| Central Africa | Rwanda | - |
| Central Africa | Rwanda | Black |
| Central Africa | Sao Tome and Principe | - |
| Central Africa | Sao Tome and Principe | Black |
| North Africa | ||
| North Africa | Algeria | - |
| North Africa | Algeria | Arab |
| North Africa | Ethiopia | - |
| North Africa | Ethiopia | Black |
| North Africa | Mali | - |
| North Africa | Mali | Black |
| North Africa | Morocco | - |
| North Africa | Morocco | Arab |
| North Africa | Morocco | Caucasoid |
| North Africa | Sudan | - |
| North Africa | Sudan | Arab |
| North Africa | Sudan | Black |
| North Africa | Sudan | Mixed |
| North Africa | Tunisia | - |
| North Africa | Tunisia | Arab |
| North Africa | Tunisia | Berber |
| South Africa | ||
| South Africa | South Africa | - |
| South Africa | South Africa | Black |
| South Africa | South Africa | Other |
| West Indies | ||
| West Indies | Cuba | - |
| West Indies | Cuba | Caucasoid |
| West Indies | Cuba | Mixed |
| West Indies | Cuba | Mulatto |
| West Indies | Jamaica | - |
| West Indies | Jamaica | Black |
| West Indies | Martinique | - |
| West Indies | Martinique | Black |
| West Indies | Trinidad and Tobago | - |
| West Indies | Trinidad and Tobago | Asian |
| North America | ||
| North America | Canada | - |
| North America | Canada | Amerindian |
| North America | Mexico | - |
| North America | Mexico | Amerindian |
| North America | Mexico | Mestizo |
| North America | United States | - |
| North America | United States | Amerindian |
| North America | United States | Asian |
| North America | United States | Austronesian |
| North America | United States | Black |
| North America | United States | Caucasoid |
| North America | United States | Hispanic |
| North America | United States | Mestizo |
| North America | United States | Polynesian |
| Central America | ||
| Central America | Costa Rica | - |
| Central America | Costa Rica | Mestizo |
| Central America | Guatemala | - |
| Central America | Guatemala | Amerindian |
| South America | ||
| South America | Argentina | - |
| South America | Argentina | Amerindian |
| South America | Argentina | Caucasoid |
| South America | Bolivia | - |
| South America | Bolivia | Amerindian |
| South America | Brazil | - |
| South America | Brazil | Amerindian |
| South America | Brazil | Caucasoid |
| South America | Brazil | Mixed |
| South America | Brazil | Mulatto |
| South America | Brazil | Other |
| South America | Chile | - |
| South America | Chile | Amerindian |
| South America | Chile | Hispanic |
| South America | Chile | Mixed |
| South America | Colombia | - |
| South America | Colombia | Amerindian |
| South America | Colombia | Black |
| South America | Colombia | Mestizo |
| South America | Ecuador | - |
| South America | Ecuador | Amerindian |
| South America | Ecuador | Black |
| South America | Paraguay | - |
| South America | Paraguay | Amerindian |
| South America | Peru | - |
| South America | Peru | Amerindian |
| South America | Peru | Mestizo |
| South America | Venezuela | - |
| South America | Venezuela | Amerindian |
| South America | Venezuela | Caucasoid |
| South America | Venezuela | Mestizo |
| South America | Venezuela | Mixed |
| Oceania | ||
| Oceania | American Samoa | - |
| Oceania | American Samoa | Polynesian |
| Oceania | Australia | - |
| Oceania | Australia | Australian Aborigines |
| Oceania | Australia | Caucasoid |
| Oceania | Chile | - |
| Oceania | Chile | Amerindian |
| Oceania | Cook Islands | - |
| Oceania | Cook Islands | Polynesian |
| Oceania | Fiji | - |
| Oceania | Fiji | Melanesian |
| Oceania | Kiribati | - |
| Oceania | Kiribati | Micronesian |
| Oceania | Nauru | - |
| Oceania | Nauru | Micronesian |
| Oceania | New Caledonia | - |
| Oceania | New Caledonia | Melanesian |
| Oceania | New Zealand | - |
| Oceania | New Zealand | Polynesian |
| Oceania | Niue | - |
| Oceania | Niue | Polynesian |
| Oceania | Papua New Guinea | - |
| Oceania | Papua New Guinea | Melanesian |
| Oceania | Samoa | - |
| Oceania | Samoa | Polynesian |
| Oceania | Tokelau | - |
| Oceania | Tokelau | Polynesian |
| Oceania | Tonga | - |
| Oceania | Tonga | Polynesian |
| Population Ethnicity |
|---|
| Amerindian |
| Arab |
| Asian |
| Australian Aborigines |
| Austronesian |
| Berber |
| Black |
| Caucasoid |
| Hispanic |
| Jew |
| Kurd |
| Melanesian |
| Mestizo |
| Micronesian |
| Mixed |
| Mulatto |
| Oriental |
| Persian |
| Polynesian |
| Siberian |