MHC-II binding predictions - Tutorial
Guidelines for selecting thresholds (cut-offs) for MHC class I and II binding predictions can be found
here.
How to obtain predictions
This website provides access to predictions of peptide binding to MHC class II molecules. The screenshot below illustrates the steps necessary to make a prediction.
Each of the steps is described in more detail below.
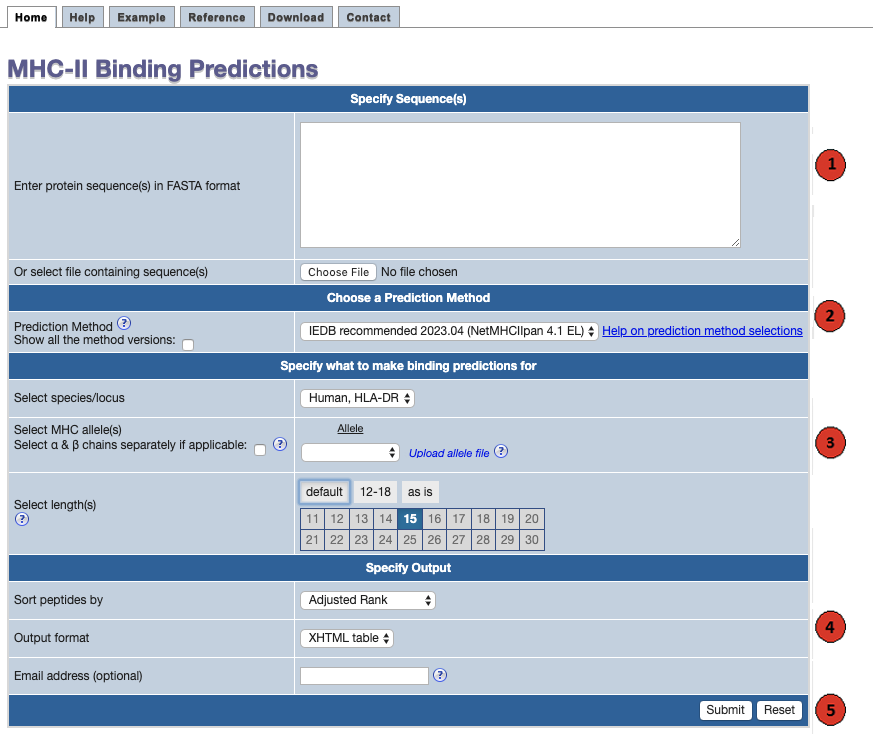
1. Specify sequences:
First specify the sequences you want to scan for binding peptides. The sequences
should either be entered directly into the textarea field labeled "Enter protein sequence(s),
or can be taken from a file that has to be uploaded using the button labeled "Browse". Please
enter no more then 200 FASTA sequences or upload file size less than or equal to 10 MB per query.
The sequences can be supplied in three different formats:
All sequences have to be amino acids specified in single letter code (
The sequences can be supplied in three different formats:
- Space separated sequences
- One continuous sequence
- FASTA format
All sequences have to be amino acids specified in single letter code (
ACDEFGHIKLMNPQRSTVWY).
2. Choose a prediction method:
The prediction method list box allows choosing between nine currently implemented
MHC class II binding prediction methods:
IEDB recommended,
Consensus method,
Combinatorial library,
NN-align-2.3 (netMHCII-2.3),
NN-align-2.2 (netMHCII-2.2),
SMM-align (netMHCII-1.1),
Sturniolo,
NetMHCIIpan-3.1,
NetMHCIIpan-3.2,
NetMHCIIpan-4.0,
NetMHCIIpan-4.1,
NetMHCIIpan-4.2, and
NetMHCIIpan-4.3.
NetMHCIIpan versions 4.0 and above include separate predictors for binding (BA) and elution (EL). While BA predictions evaluate the ability of a peptide to bind an MHC molecule, EL predictions also incorporate the likelihood that the peptide will be naturally processed and presented, which makes it more likely to be recognized as a T cell epitope. As these methods have distinct purposes and performance characteristics, as of September 2023, the IEDB has divided the recommended methods according to the intended purpose of the prediction. The weekly automated benchmarks for binding predictions consistently show that NetMHCIIpan 4.1 BA is the top-performing binding predictor. It is anticipated that the forthcoming automated benchmarks for methods trained on elution data will indicate that NetMHCIIPan 4.1 EL here is the top performer.
The Consensus approach, combining NN-align, SMM-align, CombLib and Sturniolo if any corresponding predictor is available for the molecule, otherwise NetMHCIIpan is used. The Consensus approach considers a combination of any three of the four methods, if available, where Sturniolo as a final choice. The expected predictive performances are based on large scale evaluations of the performance of the MHC class II binding predictions: a 2008 study based on over 10,000 binding affinities, a 2010 study based on over 40,000 binding affinities and a 2012 study comparing pan-specific methods. Supplementary information for evaluation of predictive tools are available for 2008 and 2010 studies.
Version method(s) used in the tool:
| Method | Version | Source |
|---|---|---|
| NetMHCIIpan EL | 4.1 | DTU |
| NetMHCIIpan BA | 4.1 | DTU |
| NetMHCIIpan EL | 4.3 | DTU |
| NetMHCIIpan BA | 4.3 | DTU |
| NetMHCIIpan EL | 4.2 | DTU |
| NetMHCIIpan BA | 4.2 | DTU |
| NetMHCIIpan EL | 4.0 | DTU |
| NetMHCIIpan BA | 4.0 | DTU |
| NetMHCIIpan | 3.2 | DTU |
| NetMHCIIpan | 3.1 | DTU |
| SMM_align (NetMHCII) * | 1.1 | DTU |
| NN_align (NetMHCII) * | 2.2 | DTU |
| NN_align (NetMHCII) * | 2.3 | DTU |
| Combinatorial library | 1.0 | LJI |
| Sturniolo | 1.0 | Sturniolo |
* NetMHCII has changed method name from SMM_align to NN_align after version 2.2.
IEDB recommended is the default prediction method selection and is updated periodically based on the availability of predictors and observe predicted performance for a given allele. Currently for peptide: MHC-II binding prediction, NetMHCIIpan 4.1 EL is used across all alleles. For prior versions of the 'IEDB recommended' method (2.22 and earlier), the Consensus method is used if available for the molecule, otherwise NetMHCIIpan is used. Of note, we fully expect the IEDB recommendation to change as we perform larger benchmarks of newly developed methods on blind datasets to determine an accurate assessment of prediction quality.NetMHCIIpan versions 4.0 and above include separate predictors for binding (BA) and elution (EL). While BA predictions evaluate the ability of a peptide to bind an MHC molecule, EL predictions also incorporate the likelihood that the peptide will be naturally processed and presented, which makes it more likely to be recognized as a T cell epitope. As these methods have distinct purposes and performance characteristics, as of September 2023, the IEDB has divided the recommended methods according to the intended purpose of the prediction. The weekly automated benchmarks for binding predictions consistently show that NetMHCIIpan 4.1 BA is the top-performing binding predictor. It is anticipated that the forthcoming automated benchmarks for methods trained on elution data will indicate that NetMHCIIPan 4.1 EL here is the top performer.
Recommended Methods used in the tool:
| IEDB Tools Version | recommended method |
|---|---|
| 2023.09 (current) | NetMHCIIPan 4.1 EL (epitope prediction) |
| NetMHCIIPan 4.1 BA (binding prediction) | |
| 2023.05 | NetMHCIIPan 4.1 EL |
| 2.22 and earlier | Consensus, if available; otherwise, NetMHCpan |
The Consensus approach, combining NN-align, SMM-align, CombLib and Sturniolo if any corresponding predictor is available for the molecule, otherwise NetMHCIIpan is used. The Consensus approach considers a combination of any three of the four methods, if available, where Sturniolo as a final choice. The expected predictive performances are based on large scale evaluations of the performance of the MHC class II binding predictions: a 2008 study based on over 10,000 binding affinities, a 2010 study based on over 40,000 binding affinities and a 2012 study comparing pan-specific methods. Supplementary information for evaluation of predictive tools are available for 2008 and 2010 studies.
3. Specify what to make predictions for:
Predictions are limited to alleles that are currently covered by specific
prediction methods. Selection of a particular prediction method will generate
a list of available alleles. User can then choose a specific allele to make
predictions or upload a file containing list of alleles. User can also choose
1 or multiple peptide length(s) for the prediction.
• Select α and β chains separately:
When the locus selected is either HLA-DP or HLA-DQ, checking the box "Select α & β chains separately if applicable" enables you to choose
alpha and beta chains separately, which makes it possible for prediction of all different chain combinations. The default (un-checked) selection list only
certain chain combinations.
• Format for the upload allele file:
File should be in simple text format containing an allele in each line (example given below).
Example:
Example:
H2-IAb
HLA-DPA1*01/DPB1*04:01
HLA-DRB1*01:01
...
Additional information regarding HLA allele frequencies and nomenclature are also provided.
HLA-DPA1*01/DPB1*04:01
HLA-DRB1*01:01
...
• Select HLA allele reference set:
When the IEDB recommended 2.22 or earlier version is selected, this box can be checked to select a reference panel of 27 alleles, as described here.
• Select "7-allele" reference set:
When the IEDB recommended 2.22 or earlier version is selected, this box can be checked to select a reference panel of 7 alleles, as described in Paul et al, 2015.
• Select length:
1 or multiple peptide length(s) can be selected for the prediction. Available length options is from 10 to 30, and the default value is 15. A list of peptides will be automatically generated for predictions by scanning input sequence(s) with user selected length(s).
• Select "asis" length option:
When the box "asis" is selected, the length(s) of input sequence(s) will be used for the prediction. But the length(s) still should be from 10 to 30, and all the other inputs will be ignored for the prediction.
4. Specify the output:
The menus in this section change how the prediction output is displayed. Using the
"Sort peptides by" listbox, the results can be presorted by the order of the peptides
in their source sequence (default) or by their predicted affinity.
To reuse the prediction results in an external program, it is possible to retrieve the predictions in a plain text format. To do this, choose "Text file" in the output format listbox.
To reuse the prediction results in an external program, it is possible to retrieve the predictions in a plain text format. To do this, choose "Text file" in the output format listbox.
• Sending the result table in a email:
Inputting your email address is recommended to ensure you could receive the result, especially for those prediction jobs which will take a long time. A email with the result table attached will send to you mail box as well as the result displayed on the web site.
One or multiple email addresses with comma separated could be accepted.
Example:
For additional information regarding how to input your email address in the mhci API, Please look at the help page here.
One or multiple email addresses with comma separated could be accepted.
Example:
youremail@example.com
email1@example.com, email2@example.com, email3@example.com ...
Please input your email address for the extremely large predictions because for these jobs we only send the result to users by email. Or download the standalone to finish these predictions locally.email1@example.com, email2@example.com, email3@example.com ...
For additional information regarding how to input your email address in the mhci API, Please look at the help page here.
5. Submit the prediction:
This one is easy. Click the submit button, and a result screen similar
to the one below should appear.
Interpreting prediction output
Below is a screenshot of a prediction output page, with three relevant sections
marked that are described in more detail below.
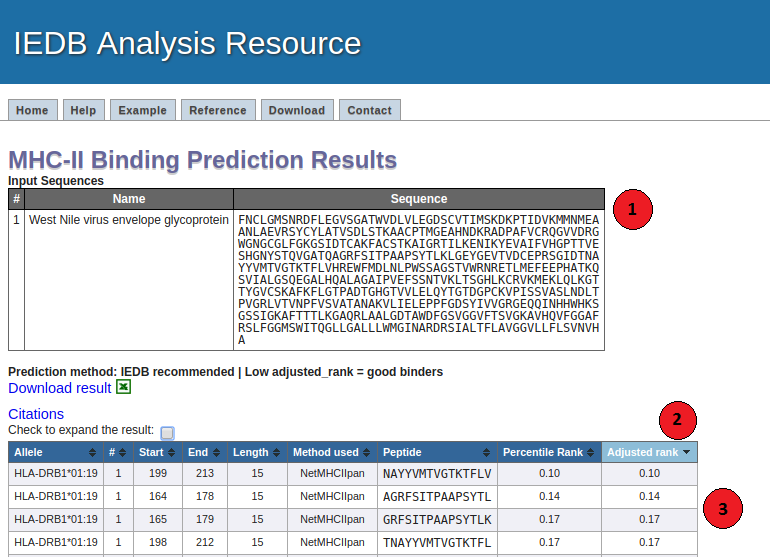
1. Input Sequences:
This table displays the sequences and their names extracted from the user input.
If no names were assigned by the user (which is only possible in FASTA format),
the sequences are numbered in their input order (sequence 1, sequence 2, ...).
2. Prediction output table:
Each row in this table corresponds to one peptide binding prediction. The columns contain the allele the
prediction was made for, the input sequence number (#), start position and end position of the peptide,
its length, the peptide, ('method used' if IEDB recommended 2.22 or earlier version is used and 'percentile rank' for both
consensus and IEDB recommended2.22 or earlier), the core sequence, the predicted score and percentile rank for combinatorial
library, SMM_align and Sturniolo. The table can be sorted by clicking on the table column headers.
3. Interpreting predicted results:
The predicted output is given in units of IC50nM for combinatorial library and SMM_align.
Therefore a lower number indicates higher affinity. As a rough guideline, peptides with IC50values
<50 nM are considered high affinity, <500 nM intermediate affinity and <5000 nM low affinity. Most known epitopes
have high or intermediate affinity. Some epitopes have low affinity, but no known T-cell epitope has an IC50
value greater than 5000.
The prediction result for Sturniolo is given as raw score. Higher score indicates higher affinity.
For each peptide, a percentile rank for each of the three methods (combinatorial library, SMM_align and Sturniolo)
is generated by comparing the peptide's score against the scores of five million random 15 mers selected from SWISSPROT
database.
And the adjusted percentile rank is the percentile rank adjusted based on the frequency of peptide lengths.
A small numbered percentile rank indicates high affinity. The median percentile rank of the three methods were
then used to generate the rank for consensus method.
The data for percentile rank was updated in the release 2.22 by recalculating the percentile data with a consistent sample peptides datasets for all the methods. If you would like to get prediction results with old ranking data, please download the previous version of the standalone to complete predictions.
Regarding the peptides selection of the latest percentile data calculation, we downloaded the "Reviewed (Swiss-Prot)" dataset from https://www.uniprot.org/downloads in FASTA format on 10/29/2018. The file contained 558,712 sequences. 555,970 of them were acceptable by tools of IEDB, and 544,147 of them had length of at least 50 aa. Then we randomly selected 10,000 of these protein sequences and further randomly selected peptides with specified lengths (8-15 for class I; 10-30 for class II) from each protein sequence. specified lengths from each sequence. Click here to download the datasets.
Regarding the peptides selection of the latest percentile data calculation, we downloaded the "Reviewed (Swiss-Prot)" dataset from https://www.uniprot.org/downloads in FASTA format on 10/29/2018. The file contained 558,712 sequences. 555,970 of them were acceptable by tools of IEDB, and 544,147 of them had length of at least 50 aa. Then we randomly selected 10,000 of these protein sequences and further randomly selected peptides with specified lengths (8-15 for class I; 10-30 for class II) from each protein sequence. specified lengths from each sequence. Click here to download the datasets.
4. Default prediction output table:
By default prediction result is collapsed to show only the Percentile Rank and Adjusted Rank when the Consensus method is
used. The table can be expanded to display the individual score of different methods used by checking box above result
table.
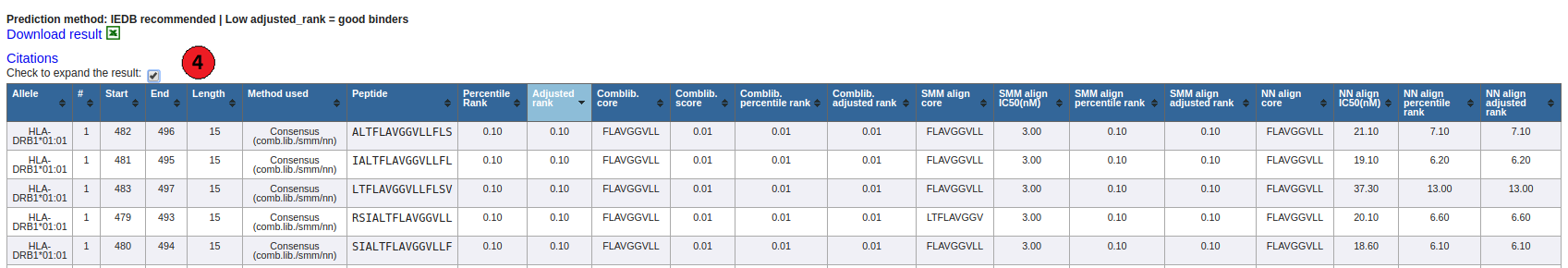

5. Predicted results:
NetMHCIIpan method is used when Consensus and other methods such as SMM_align, NN_align, COMBLIB and/or Sturniolo are
not available for a particular allele. However, if only one or two of these methods are available, NetMHCIIpan is
used as second or third method.

