* This tool has been developed by Gustavo Fioravanti Vieira's group and has been deployed to the IEDB-AR servers with minimal modification by the IEDB team.
DockTope is a web-based tool, based on D1-EM-D2 approach, intended to allow the pMHC-I modeling.
Some applications allowed from the pMHC-I construction include:
- The study of the pMHC-I tridimensional structure
- Epitope-MHC-I interaction patterns
- Cross-reactivity assessment
- Molecular dynamics studies
This site provides a tool to construct pMHC-I structures for the following alleles: HLA-A*02:01, HLA-B*27:05,
H2-Db and H2-Kb.
Latest MHC-I alleles included
- HLA-A*02:01 (9-mer epitopes)
- HLA-B*27:05 (9-mer epitopes)
- H2-Db (9-mer /10-mer epitopes)
- H2-Kb (8-mer epitopes)
Getting started
See the FAQ page to get started.
Disclaimer
1) It should be noted that our team employed all efforts, based on experimental data, to develop this tool. Though,
the use of the modeled structures, and also all the files generated from this web tool, should be carefully examined
by the user.
2) All files will be available for download during a period of 2 weeks, after the conclusion of the job. After this
period, all files will be removed from our server.
D1-EM-D2 approach pipeline
The D1-EM-D2 approach, used as basis for the DockTope, could be visualized through the following flowchart:
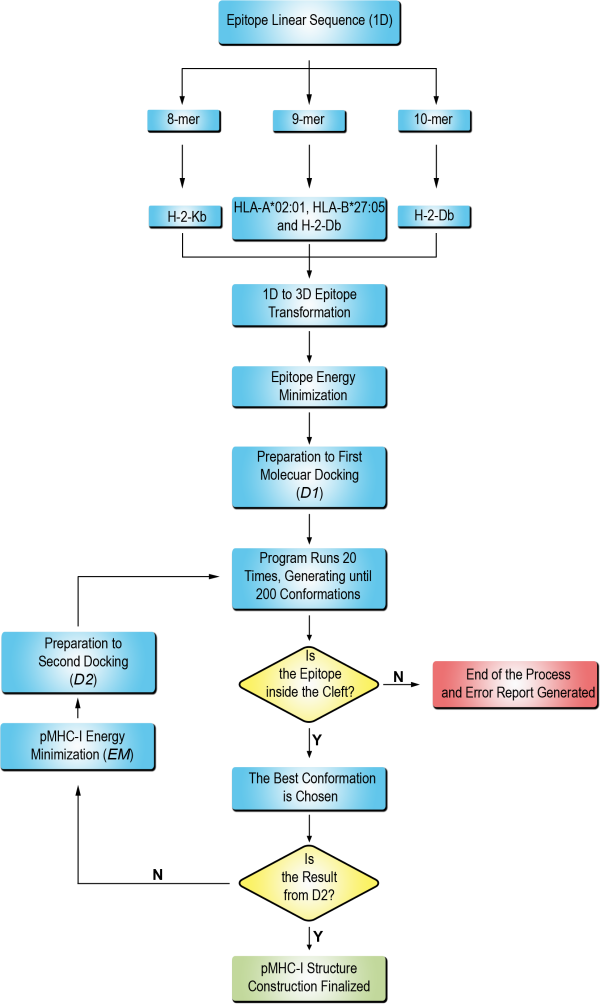
Briefly, the technique occurs consecutively throughout the following steps:
-
The epitope sequence provided by the user is modeled according to the MHC-I epitope pattern.
-
A molecular docking (D1) is performed using Autodock Vina program between the MHC-I and the epitope provided.
-
An energy minimization step (EM) is performed to correct possible steric clashes between the epitope and the MHC-I
-
A second molecular docking (D2) is performed in order to refine the structure.
Release notes
There are no release notes so far.
Acknowledgements
We would like to acknowledge all the people involved to give shape and reliability for the development of this tool. A special acknowledgement to:
-
CESUP (Centro de Supercomputação da Universidade Federal do Rio Grande do Sul) for the technical support and for provides adequate facilities to run the jobs.
-
Dr. José Artur Bogo Chies, PI of Immunogenetics Lab (UFRGS), for immunological discussions and for provides material support for our research.
Useful links
- GROMACS: http://www.gromacs.org
- AutoDock Tools: http://autodock.scripps.edu/resources/adt
- AutoDock Vina: http://vina.scripps.edu/
- PyMOL: http://www.pymol.org/
- Chimera: http://www.cgl.ucsf.edu/chimera/
Articles
RIGO, M. M. ; ANTUNES, D. A. ; FREITAS, M. V. ; MENDES, M. F. A. ; MEIRA, L. ; SINIGAGLIA, M. ; VIEIRA, G. F. Docktope: a Web-based tool for automated pMHC-I modeling. Scientific Reports, v.5, online, 2015. doi:10.1038/srep18413
SINIGAGLIA, M. ; ANTUNES, D. A. ; RIGO, M. M. ; CHIES, J. A. B. ; VIEIRA, G. F. CrossTope: a curate repository of 3D structures of immunogenic peptide:MHC complexes. DATABASE-OXFORD, v. 2013, p. bat002-bat002, 2013.
ANTUNES, D. A. ; VIEIRA, G. F. ; RIGO, M. M. ; CIBULSKI, S. P. ; SINIGAGLIA, M. ; CHIES, J. A. B. Structural Allele-Specific Patterns Adopted by Epitopes in the MHC-I Cleft and Reconstruction of MHC:peptide Complexes to Cross-Reactivity Assessment. Plos One, v. 5, p. e10353, 2010.
FÜLBER, C.C.; ANTUNES, D.A.; RIGO, M.M.; CHIES, J. A.B.; SINIGAGLIA, M.; VIEIRA, G.F. Reconstruction of MHC Alleles by Cross Modeling and Structural Assessment. In: BIOCOMP'10 USA, 2010, Las Vegas. International Conference on Bioinformatics and Computational Biology, 2010.
RIGO, M. M. ; ANTUNES, D. A. ; VIEIRA, G. F. ; CHIES, J. A. B. MHC: Peptide Analysis: Implications on the Immunogenicity of Hantaviruses N protein. Lecture Notes in Computer Science, v. 5676, p. 160-163, 2009.
