1. Introduction
MHC class I antigen processing consists of multiple steps that result in the
presentation of MHC bound peptides that can be recognized as T cell epitopes.
Many of the pathway steps can be predicted using computational methods, but one
is often neglected: mRNA expression of the epitope source proteins. We improve
epitope prediction by taking into account both peptide-MHC binding affinities
and expression levels of the peptide’s source protein. Specifically, we utilized
biophysical principles and existing MHC binding prediction tools in concert with experimental
RNA expression values to derive a function that estimates the likelihood of a peptide
being presented on a given MHC class I molecule. Our combined model of Antigen eXpression
based Epitope Likelihood-Function (AXEL-F) outperformed predictions based only on binding
or based only on antigen expression for discriminating eluted ligands from random background
peptides as well as in predicting neoantigens that are recognized by T cells.
2. Input
Axel-F can accept either CSV file or FASTA formatted file. Note that when providing CSV file
as an input for Axel-F, it must be in a valid CSV format. A valid CSV file will contain a header
and have no missing data or empty cell in each row. As for FASTA formatted input, it should be a
valid format that agrees with the
NIH standards.
Obtain predictions using CSV formatted inputs
There are multiple ways of what to include inside the CSV file. The simplest form of data
you can provide to Axel-F is a CSV file containing peptide sequences only. Depending on
available data you have, you may include allele names, tpm values, and/or gene names at your choice.

Example 1.
Contains peptide sequence only.
(Simplest form of data.)
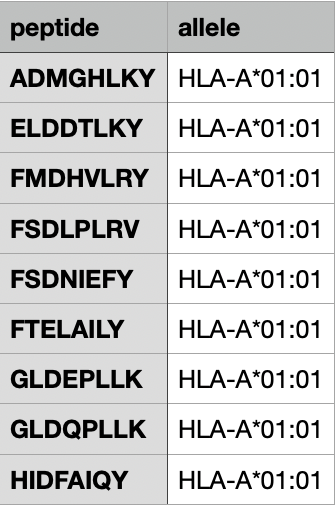
Example 2.
Contains peptide sequence and allele names.
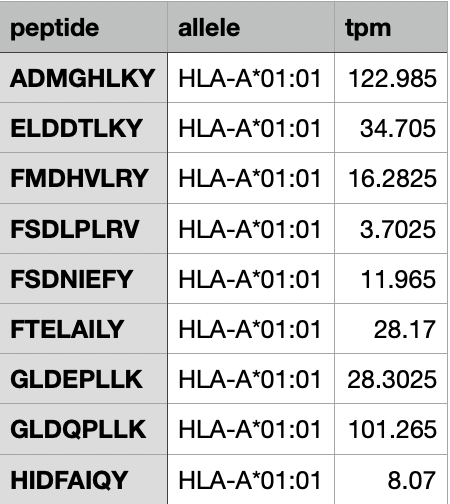
Example 3.
Contains peptide sequence, allele names, and TPM value.
(Using custom TPM values.)
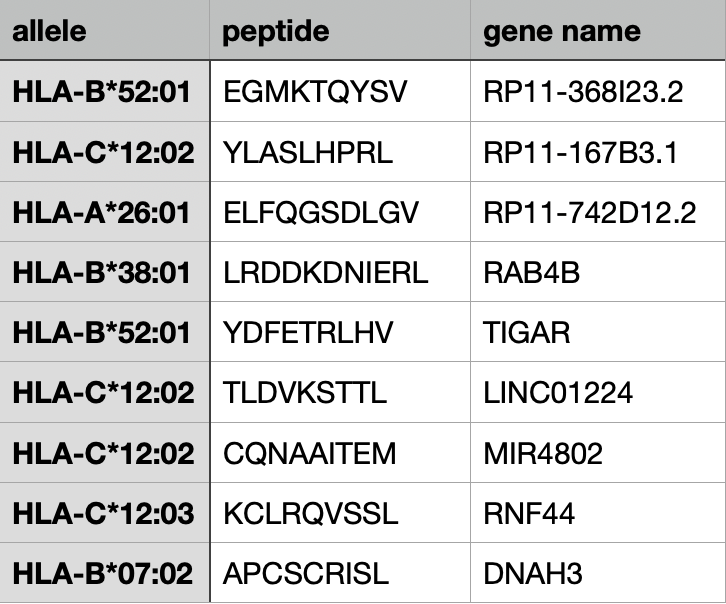
Example 4.
Contains peptide sequence, allele names, gene names.
(Using TPM values from TCGA data.)
Please note that these are CSV formatted file, thus TSV format would not work. If you would like to
simply create CSV formatted file on a text editor or directly on the form, please have each column
separated by comma.
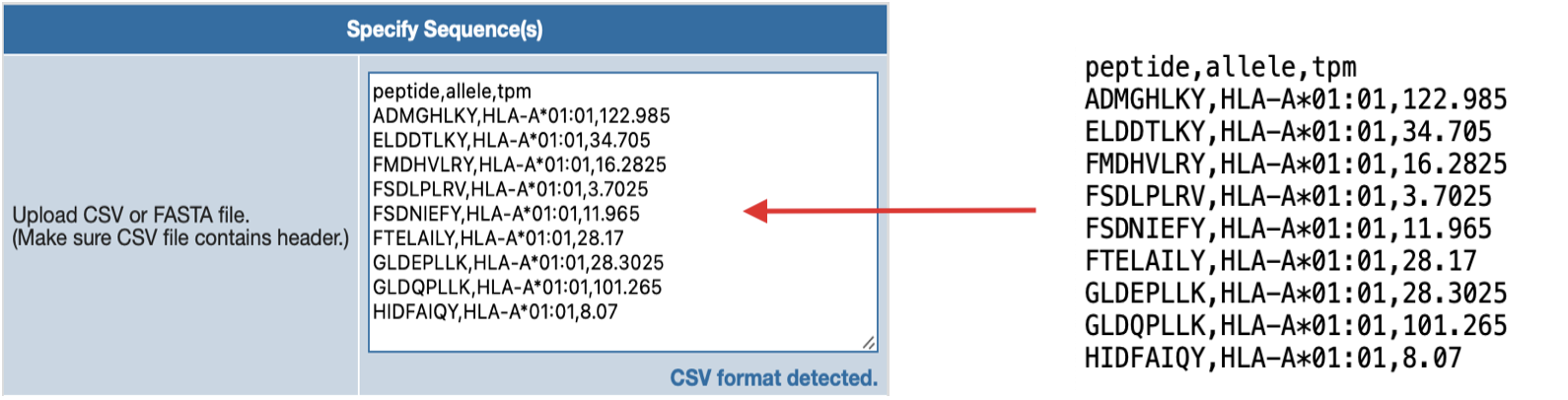
CSV Format Example In Plain Text
If you filling out data manually on text file/editor or directly on the AXEL-F form,
make sure you separate each data with comma if you have more than one column.
Because Axel-F requires allele and TPM value, you must specify them on the form if not already
included in the CSV file. Let's take Example 1, which includes peptide sequences only. Axel-F would require
you to specify allele and TPM value on the form. If Example 2 was provided, you don't need to specify allele
on the form, but you still do need to provide TPM value. If Example 3 was provided, you don't have to
specify anything since required information (Peptide sequence, allele names, TPM values) were all provided
inside the CSV file.
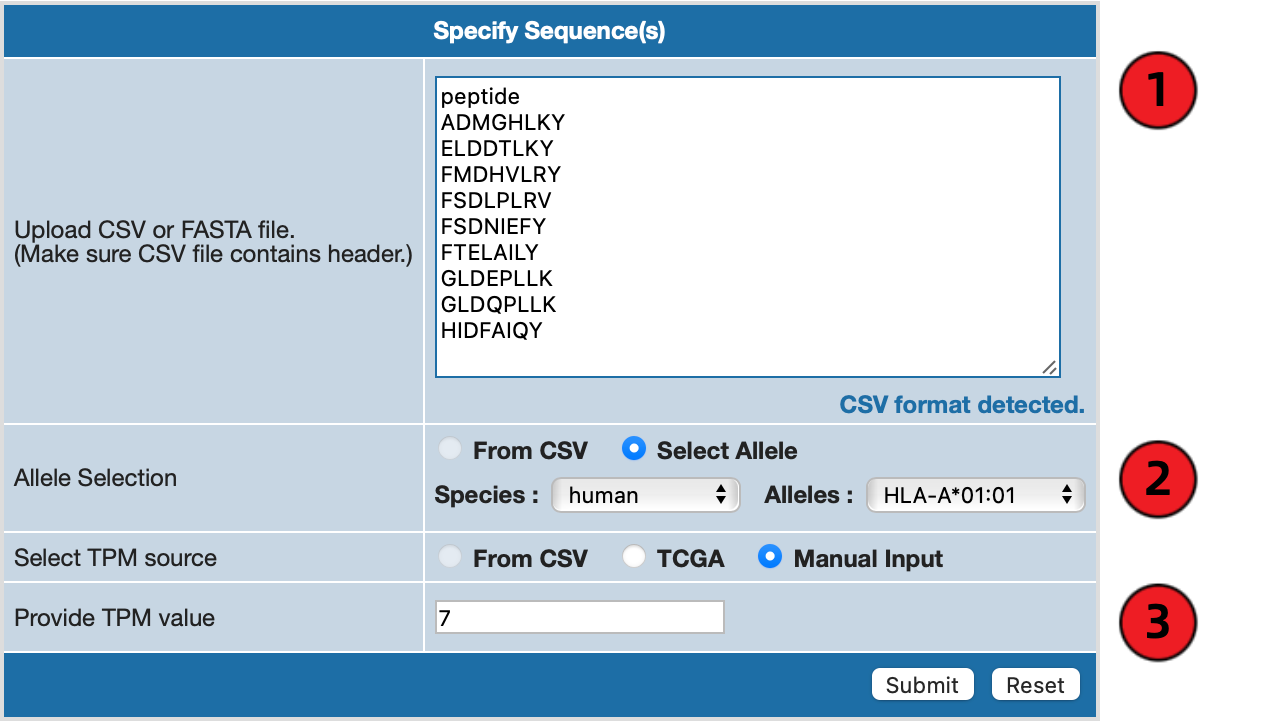
1. Specify Sequences
Using Example 1, you can either manually write out the sequences in the textarea with the header "peptide" included,
or drag and drop a CSV file containing peptide sequences onto the textarea. If you want to include any other data,
make sure you include proper header on top, such as
"allele",
"tpm", or
"gene name".
2. Specify Allele
Axel-F requires at least one allele to be specified in order to run the prediction. Unless you provided "allele"
column in the CSV file, please select species of interest and appropriate allele from the dropdown menu on the form.
3. Specify TPM Value Manually
If you have a custom TPM value in mind, please enter the value in the textfield or include it in the CSV file.
Otherwise, you may specify TPM value in the form. Also note that TPM value must be an integer or decimal value.
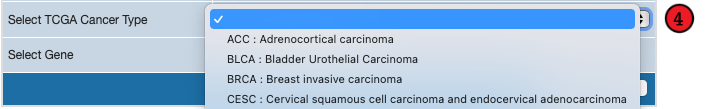
4. Specify TPM Value through TCGA
If you would like to get TPM value derived from TCGA data instead, click on "TCGA" button for "Select TPM source"
and specify both
cancer type and
gene name. If the CSV file already contains list of gene names,
you only need to specify cancer type on the form.
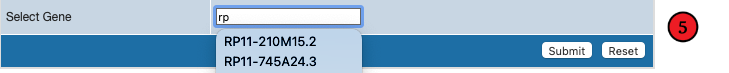
As you type in gene name, it will also autosuggest or recommend available genes.
Obtain predictions using FASTA formatted inputs
FASTA formatted input is a lot more direct compared to CSV file as there really only one type of format that Axel-F
can accept. A valid FASTA format is single-line description starting with ">" character, followed by lines of sequence data.
** Note : Currently Axel-F can accept only one FASTA sequence at a time.

If you fill the textbox with FASTA formatted data, AXEL-F form will detect that it's a FASTA format and will
change available options. Compared to CSV format options, you can see from the following image that the form
now has a length option added to it.
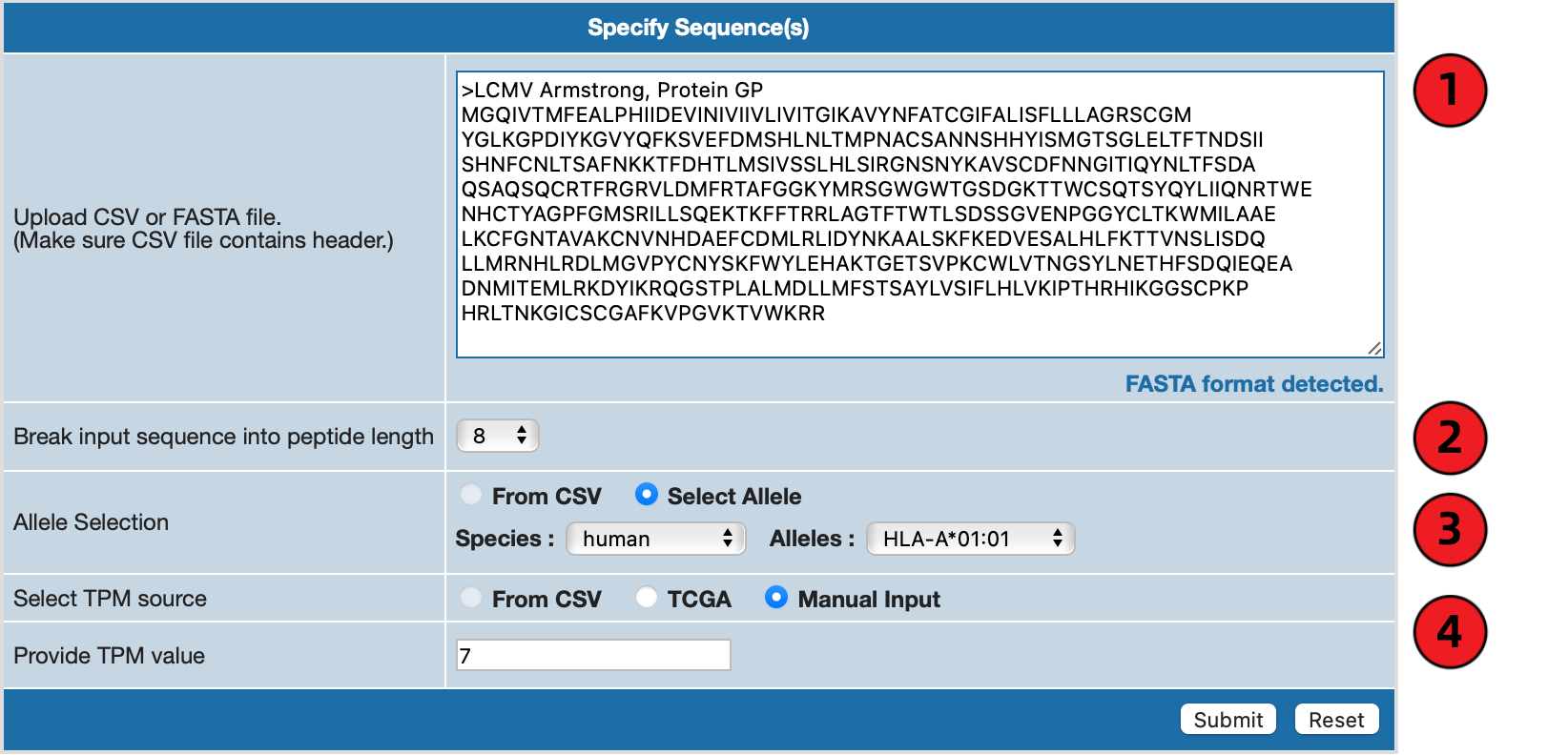
1. Specify Sequences
You can either manually write out the sequences in the textarea in proper FASTA format (starting with signle-line
description starting with ">" character) or drag and drop an existing FASTA file onto the textarea.
2. Specify Length
Unlike with CSV examples, predictions are limited to one specific binding length. Selecting one length will be applied
to all peptides from the FASTA sequence.
3. Specify Allele
Axel-F requires at least one allele to be specified in order to run the prediction.
Please select species of interest and appropriate allele from the dropdown menu on the form.
4. Specify TPM Value Manually or From TCGA
If you have a custom TPM value in mind, please enter number in the textbox.
Also note that TPM value must be an integer or decimal value.
If you would like to get TPM value derived from TCGA data instead, click on "TCGA" button for "Select TPM source"
and specify both
cancer type and
gene name.
3. Interpreting prediction output
Continuing with
Example 1 from CSV input, after submitting the form, you should get the following results.
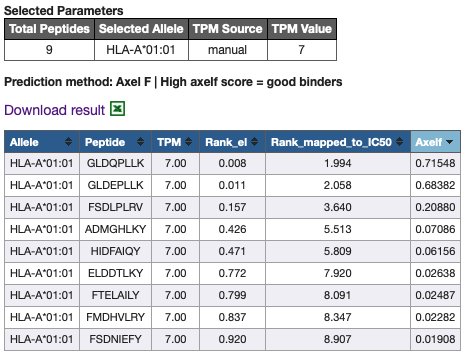
Rank EL values are directly from the neural networks that NetMHCpan uses. Because these numbers are abstract and cannot
be directly used in biological context of Axelf, Rank EL values are translated to IC50 values by comparing the
percentile ranks of the two metrics in Trolle dataset, then using interpolation function to map each percentile ranks
to corresponding IC50 value. The resulting values are stored in
Rank_mapped_to_IC50 column.
The same interpolation function based in the
Trolle dataset
was used to calculate AXEL-F scores.
AXEL-F scores estimates the likelihood of a peptide being presented on HLA and being an epitope.











