Please refer to the manuscripts provided at the Reference tab for more details.
Purpose of the tool:
Input data:
Result page:
Output parameters (Complete/Concise results):
How to interpret results:
How the tool works:
Special Considerations:
Note: Please refer the manuscript for more details or contact us at help@iedb.org if you have further questions. Please include the "Job Id" in your correspondence if you have any.
Purpose of the tool:
RATE (Restrictor Analysis Tool for Epitopes) is an automated method that can infer HLA restriction for a set of given epitopes from large datasets of T cell responses in HLA typed subjects.
The tool takes two data files, one containing the alleles expressed by the subjects and the other containing the response of the peptides in the subjects.
The tool calculates the odds ratio and estimates its significance using Fisher's exact test. It also calculates a parameter called relative frequency similar to odds ratio.
The tool was developed with a focus on class II alleles but can also be applied to class I alleles.
Input data:
- Allele typing data of the subjects:
- First row: Donor/subject names (e.g., Donor-1, Donor-2, etc.)
- Subsequent rows: Each row represents a different HLA locus (e.g., DRB1, DRB3, DRB4, DRB5, DQA1, DQB1, DPA1, DPB1)
- Each cell: Contains the specific allele for that donor at that locus (e.g., DRB1*03:01, DQA1*02:01)
- Missing data: Represent unknown allele types as 'n/a'
- HLA prefix: Both 'DRB1*03:01' and 'HLA-DRB1*03:01' formats are supported
- Response data:
- First row: Column headers: Peptide #, Peptide_ID, Peptide_Seq, followed by donor names
- Subsequent rows: Each row represents a different peptide
- Columns 1-3: Peptide #, Peptide_ID, Peptide_Seq
- Remaining columns: Response values for each donor
- Response values: Can be either absolute values (e.g., 98, 0, 120) or binary values (1/0)
- Missing data: Represent unknown response values as 'n/a'
- Absolute values: Raw assay measurements (e.g., cytokine levels, proliferation indices) - the tool will apply the cutoff threshold to convert these to binary
- Binary values: Pre-processed 1/0 values indicating positive/negative responses
- Cutoff for response to be considered positive:
- For absolute values: Provide the desired cutoff threshold (e.g., 20.0 for the sample files)
- For binary values: Provide 1.0 as the cutoff (all non-zero values will be considered positive)
The allele typing data for each subject should be provided as a tab-delimited plain text file. This can be generated by copying data from a spreadsheet to a plain text file.
Format requirements:
A sample allele data input file can be downloaded from here.
The response data should be provided as a tab-delimited plain text file containing the response data for each peptide in each of the subjects.
Format requirements:
A sample response data input file can be downloaded from here.
This is the threshold value used to determine if a peptide had a positive response in a donor or not.
Usage guidelines:
Please provide 20.0 as cutoff for the sample files given above.
- Please make sure that the number and names of patients/donors are exactly the same in both input files
- Donor names must match exactly between the two files (case-sensitive)
- Both files should have the same number of donor columns
- Empty cells or missing data should be represented as 'n/a'
The result page shows:
Data Processing Details:- Details of input data
- Reformatted allele data showing the list of alleles expressed by the subjects & binary values of their presence/absence in each donor.
- Reformatted response data showing which all peptides gave positive response in each donor as binary values. This will be same as the input response data if it is provided as binary values. Note: When the input response data contains absolute values (e.g., assay measurements), the tool automatically converts these to binary values (1/0) based on the specified cutoff threshold. All response values below the threshold are set to 0, while values at or above the threshold are set to 1.
- Details of RATE results
- Link to download RATE results (i.e. the selected HLA allele restrictions)
- Link to download the complete report that shows the analysis results for all peptide-HLA allele combinations. For e.g. if there are 25 peptides and 100 alleles expressed by the subjects, results for 25 x 100 = 2500 peptide-allele combinations are shown.
- Job Id: Please let us know this number if you have any question regarding any task that you did using the tool.
Response Data Thresholding:
When response data contains absolute values (e.g., cytokine measurements, proliferation indices, etc.), the tool automatically applies the user-specified cutoff threshold to convert these values to binary format:
When response data contains absolute values (e.g., cytokine measurements, proliferation indices, etc.), the tool automatically applies the user-specified cutoff threshold to convert these values to binary format:
- Values ≥ cutoff threshold: Converted to 1 (positive response)
- Values < cutoff threshold: Converted to 0 (negative response)
- Missing values (n/a): Remain as 'n/a' (not included in statistical calculations)
Output parameters (Complete/Concise results):
- A+R+: No. of subjects expressing the allele and have a positive response
- A-R+: No. of subjects that do not express the allele but have a positive response
- A+R-: No. of subjects expressing the allele but do not have a positive response
- A-R-: No. of subjects that do not express the allele and do not have a positive response
- No. of subjects: Total no. of subjects for the peptide-allele combination
- Response n/a: No. of subjects where response value is not available
- Odds ratio (OR): OR indicates the strength of association between expression of a specific allele and detection of positive immune response. An OR > 1 indicates a positive association. OR is calculated as:
- Relative frequency (RF): RF can be used in place of OR where OR becomes “infinity” due to division by zero (when none of the donors that do not express the allele have a positive response, i.e. A-R+ = 0). RF is calculated as:
- P-value: P-value obtained from Fisher's exact test can be used as a relative ranking which restriction has most statistical support.
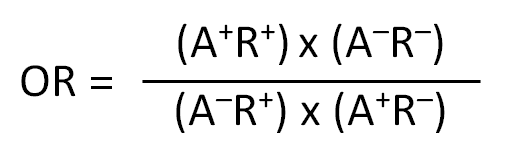
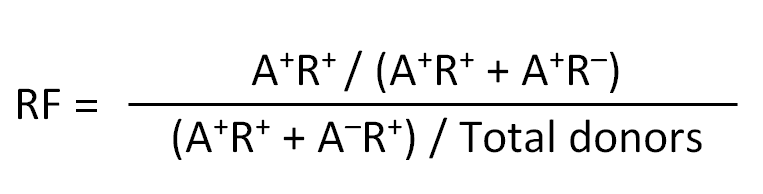
The peptide-allele combinations listed in the "RATE results" file are the inferred HLA restrictions.
The "complete report" shows the analysis results for all peptide-allele combinations.
The "complete report" shows the analysis results for all peptide-allele combinations.
How the tool works:
For each peptide-allele combination, RATE generates a matrix tabulating the number of positive versus negative responders amongst the subjects expressing that particular HLA type,
as well as the number of positive and negative responders among the subjects NOT expressing that HLA type.
Next, for each epitope RATE calculates the OR and RF corresponding to each of the HLA molecules following classical formulas, as described above.
The Fisher’s exact test is used to calculate the significance (p-value) associated with each peptide-allele combination.
Special Considerations:
DRB3/4/5 Alleles:
The tool handles DRB3, DRB4, and DRB5 alleles with special consideration, supporting both 'DRB3*02:02' and 'HLA-DRB3*02:02' formats. These alleles are processed as synonyms regardless of the HLA prefix presence.
The tool handles DRB3, DRB4, and DRB5 alleles with special consideration, supporting both 'DRB3*02:02' and 'HLA-DRB3*02:02' formats. These alleles are processed as synonyms regardless of the HLA prefix presence.
Note: Please refer the manuscript for more details or contact us at help@iedb.org if you have further questions. Please include the "Job Id" in your correspondence if you have any.
